How can we pick up DNA's faintest signals?
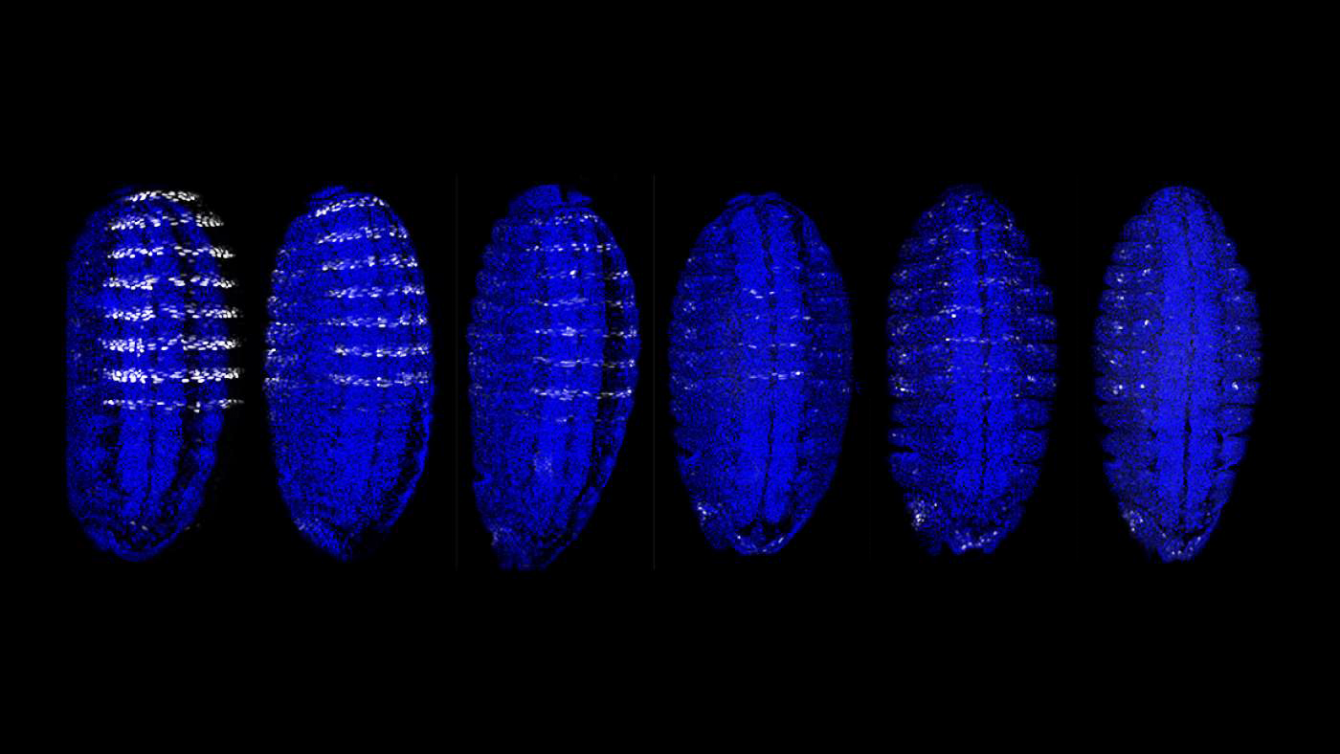
The genome is the body’s instruction manual. Its raw information—in the form of DNA—determines everything from whether an animal walks on four legs or two to one’s potential risk for disease. But this manual is written in the language of biology, so making sense of all that it encodes has proven challenging.
Columbia researchers, in a team led by the Zuckerman Institute's Richard Mann and Professor Harmen Bussemaker of the Departments of Biological Sciences and Systems Biology, have developed a computational tool, an algorithm named No Read Left Behind (NRLB), that shines a light on the most hard-to-translate segments. With this tool in hand, scientists can get closer to understanding how DNA guides everything from growth and development to aging and disease.
“Diseases such as schizophrenia, Parkinson’s, and autism have been mapped to particular DNA regions that do not appear to have a clear function," said Mann. "With NRLB, scientists could potentially piece together how transcription factors bind to and activate those regions. This will be critical for finding ways to manipulate that activity to one day reduce one’s risk of disease.” Learn more.
Make Your Commitment Today