Will new methods accelerate the discovery of cancer treatments?
A new technique for genomic sequencing promises a path to fast and inexpensive identification of the most effective drugs to combat a particular patient’s tumors. The new method combines an innovation in sequencing known as PLATE-Seq with an algorithm that matches drugs to the tumor after sequencing.
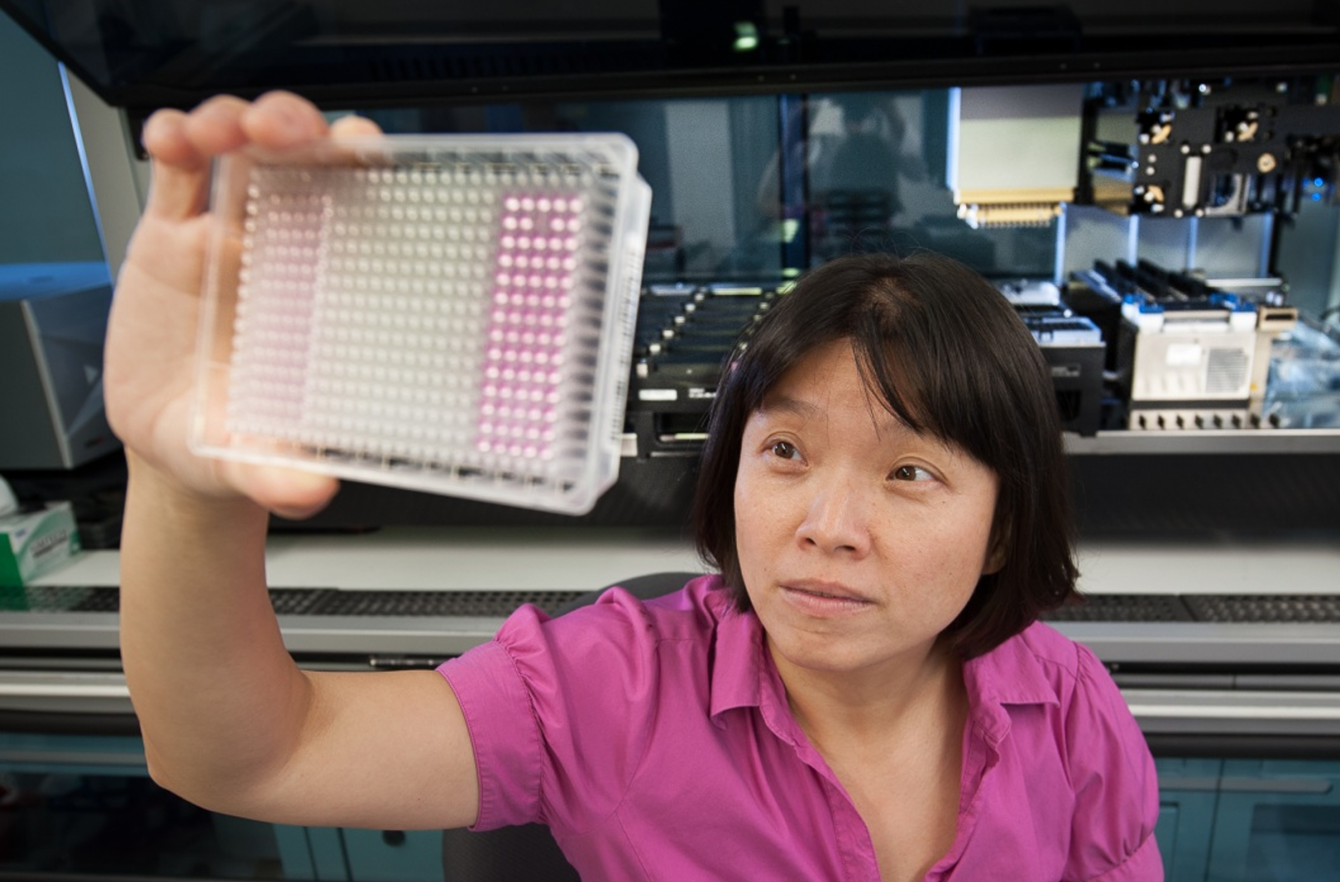
Sequencing is a process that reveals the genes that are being actively expressed, making it possible to identify potential drug targets. In conventional sequencing, researchers prepare a plate containing 96 wells of different RNA fragments from the same tumor. Every fragment is analyzed separately through the application of expensive reagents.
Peter Sims, assistant professor of systems biology, and his lab developed PLATE-Seq, a platform that reduces the standard 96-well sample plate to a single test tube. By pooling the samples, PLATE-Seq makes it possible to analyze them simultaneously. This efficiency reduces the cost of genome-wide screening from around $400 per sample to approximately $25 per sample. In a second step, the sequencing results are evaluated for potential reactivity to drugs using an algorithm developed by Andrea Califano, the Clyde and Helen Wu Professor of Chemical Systems Biology.
Using PLATE-Seq, Sims and Califano recently launched cancer treatment studies for a wide variety of tumors. They have already identified nine tumor types that show responses to existing FDA-approved drugs and have started clinical trials to evaluate these drugs as potential treatments. Learn more.
Make Your Commitment Today